Chapter 2. Search problems
“Search” is such a broad term that this entire book could be called “Classic Search Problems in Swift.” This chapter is about core search algorithms that every programmer should know. It does not claim to be comprehensive, despite the declaratory title.
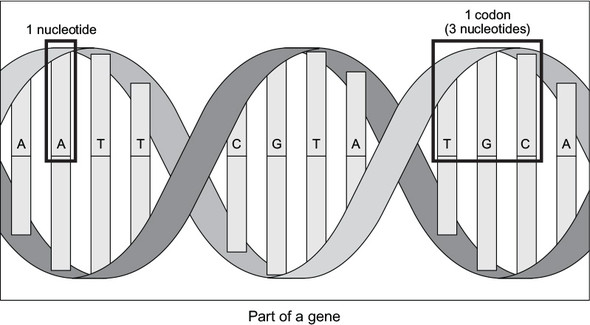
Genes are commonly represented in computer software as a sequence of the characters A, C, G, and T. Each letter represents a nucleotide, and the combination of three nucleotides is called a codon. This is illustrated in figure 2.1. A codon codes for a specific amino acid that together with other amino acids can form a protein. A classic task in bioinformatics software is to find a particular codon within a gene.
Figure 2.1. A nucleotide is represented by one of the letters A, C, G, and T. A codon is composed of three nucleotides, and a gene is composed of multiple codons.
We can represent a nucleotide as a simple enum with four cases.
Nucleotide needs to implement the Comparable interface so that Nucleotides can be put in order. An entity that implements Comparable must override the < operator. This can be done either as a freestanding function, or as a static method inside the Comparable entity.
Here, we implement < as a freestanding function by comparing one Nucleotide’s raw Character value against another’s. Character has a built-in alphabetical ordering.