appendix-c
appendix C Building knowledge graphs from structured sources
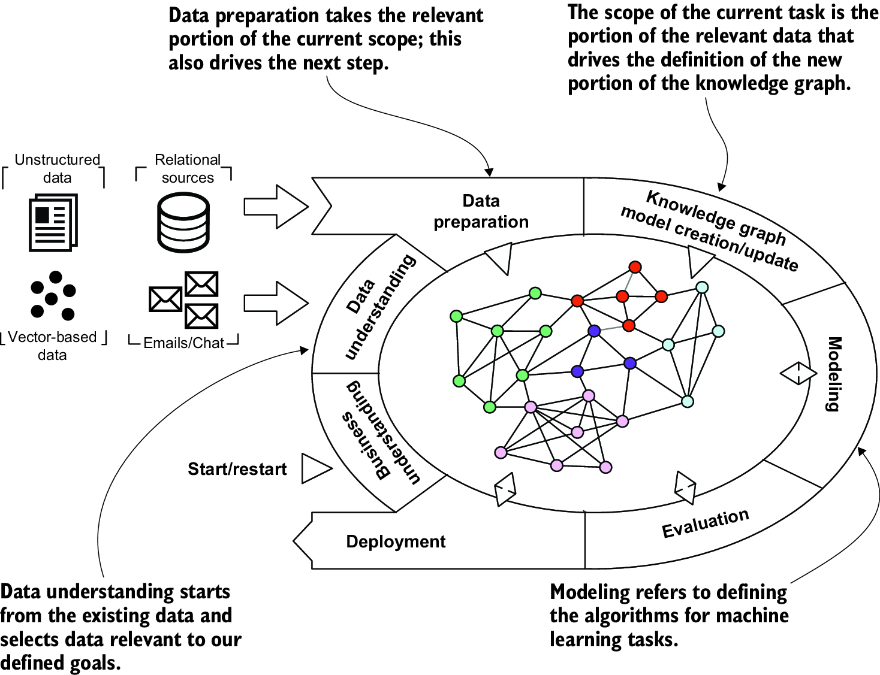
This appendix teaches you how to build your own knowledge graph (KG) from structured data sources. As we do in several places in the book, we focus on a biomedical use case: here, the detection of microRNA disease associations. Figure C.1 shows the core of the KG we’ll build. For this project, we will use CRISP-DM model introduced in chapter 2 (see figure C.2).
Figure C.1 An example of the relationship between a disease (celiac disease) and microRNAs
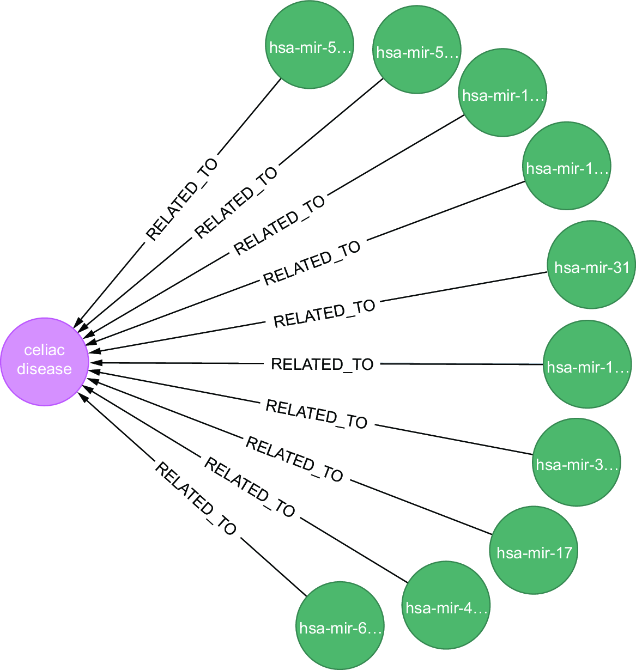
Figure C.2 CRISP-DM model adapted to KGs
C.1 MicroRNA–disease association: Warmup
MicroRNA–disease association is a very relevant use case for KGs in the biomedical space. In this section, we provide a brief biological description of what microRNAs are and how they are connected to diseases. We then outline the business goals and the data available to us for achieving those goals.